The SAMSON School is split into two independent parts, and you can register to either part, or both of them:
- The Users program from Monday at 10:30 am to Wednesday at 2pm
- The Developers program from Wednesday at 10:30 am to Friday at 2pm
- The Full program from Monday at 10:30 am to Friday at 2pm
Day 1 – Monday, June 19
Beginning of the Users program
- 10:30 am – 12:30pm: Arrival
- 12:30 pm – 2:00 pm: Lunch
- 2:00 pm – 3:30 pm: Welcome – Introduction and installation of SAMSON - Show (all) - Hide (all) - View tutorial

We introduce the Users program of the SAMSON School and SAMSON, the software platform for computational nanoscience. We highlight the need for an integrated platform able to combine diverse domains of nanoscience, which led to SAMSON's open architecture and the creation of the SAMSON Connect online platform. We introduce SAMSON Connect, the online platform used to distribute SAMSON and SAMSON Elements, the modules for SAMSON, and we install SAMSON and default SAMSON Elements.
- 3:30 pm – 4:00 pm: Coffee break
- 4:00 pm – 5:00 pm: Basics I – Interface, documents, preferences and cameras - Show (all) - Hide (all)
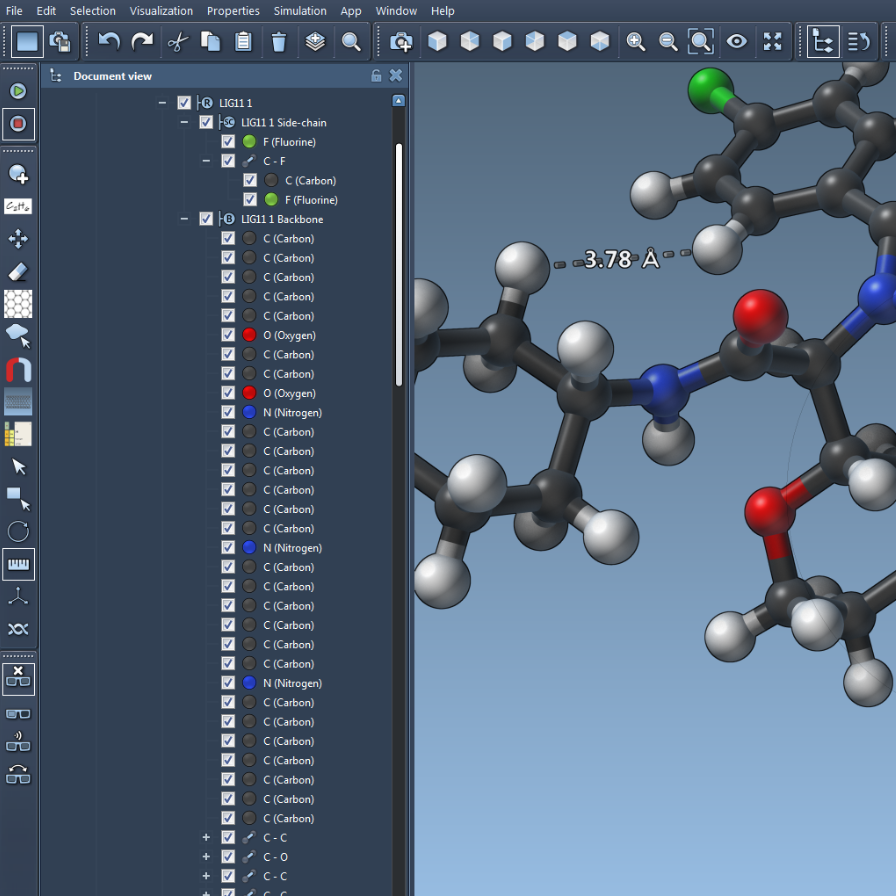
We introduce the SAMSON interface design, with its menus and toolbars for apps, editors, parsers, visualization, simulation, etc. We detail the structure of SAMSON documents and describe the SAMSON data graph, including data graph nodes for models, simulators, labels, conformations, groups, etc. We explain how to control the document history and undo / redo commands, and manage the document view. We explain how to modify preferences and control the camera.
- 5:00 pm – 6:00 pm: Basics II – Editors, apps, context menus, groups and conformations - Show (all) - Hide (all)
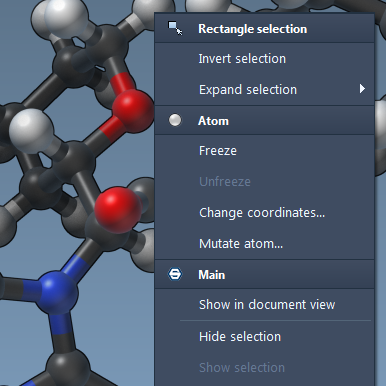
We demonstrate essential editors to select, create and modify basic molecular structures. We explore the concept of SAMSON apps and demonstrate a simple app to download PDB models from the Protein Data Bank. We explore context menus and groups, and give an overview of interactive simulation in SAMSON. Finally, we introduce conformation nodes in SAMSON to store and retrieve molecular conformations.
- 7:00 pm: Dinner
Day 2 – Tuesday, June 20
- 7:30 am – 8:30 am: Breakfast
- 8:30 am – 10:00 am: Producing publication-quality images and animations - Show (all) - Hide (all) - View tutorial

We explore how SAMSON can be used to generate publication-quality images and animations, thanks to visual models and color schemes, and fine control of rendering options and effects, including interactive ambient occlusion, depth-of-field rendering, silhouettes, anti-aliasing, multisampling, etc. We show how to setup SAMSON to capture snapshots of the viewport.
- 10:00 am – 10:30 am: Coffee break
- 10:30 am – 11:30 am: Selecting and modeling with the Node Specification Language - Show (all) - Hide (all)

We describe SAMSON's powerful Node Specification Language (NSL), and demonstrate how it can be used to perform advanced selections and modeling tasks, such as finding a binding site in a protein, building a nanopore, etc.
- 11:30 am – 12:30 pm: Simulating small molecules, graphene and proteins - Show (all) - Hide (all) - View tutorial
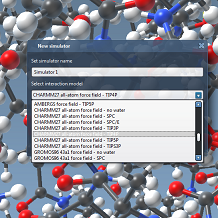
We demonstrate how SAMSON can be used to perform interactive simulation of various systems, including small molecules with the Universal Force Field (UFF), graphene and nanotubes with the Brenner force field, and proteins with GROMACS force fields. We show how interactive minimization helps produce realistic structures. We discuss the choice of state updaters.
- 12:30 pm – 2:00 pm: Lunch
- 2:00 pm – 2:30 pm: Calculating non-linear normal modes - Show (all) - Hide (all) - View tutorial
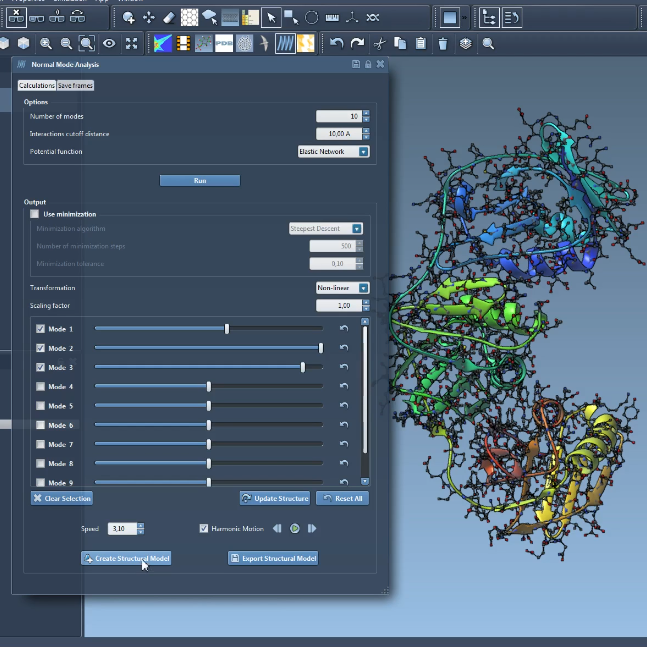
We demonstrate the SAMSON app which makes it possible to compute non-linear normal modes very efficiently. We show how they help produce large realistic deformations of protein structures, and demonstrate how to generate and export trajectories and snapshots.
- 2:30 pm – 3:00 pm: Aligning sequences with MUSCLE – Interactive Ramachandran plots - Show (all) - Hide (all) - View tutorial
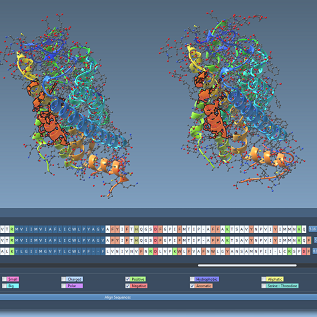
We show how to use the SAMSON app that integrates the MUSCLE tool for multiple sequence alignement, and we demonstrate the SAMSON app for interactive Ramachandran plots, which lets users directly edit the Ramachandran plot to modify the protein structure.
- 3:00 pm – 3:30 pm: Analyzing small-angle X-ray scattering data - Show (all) - Hide (all)
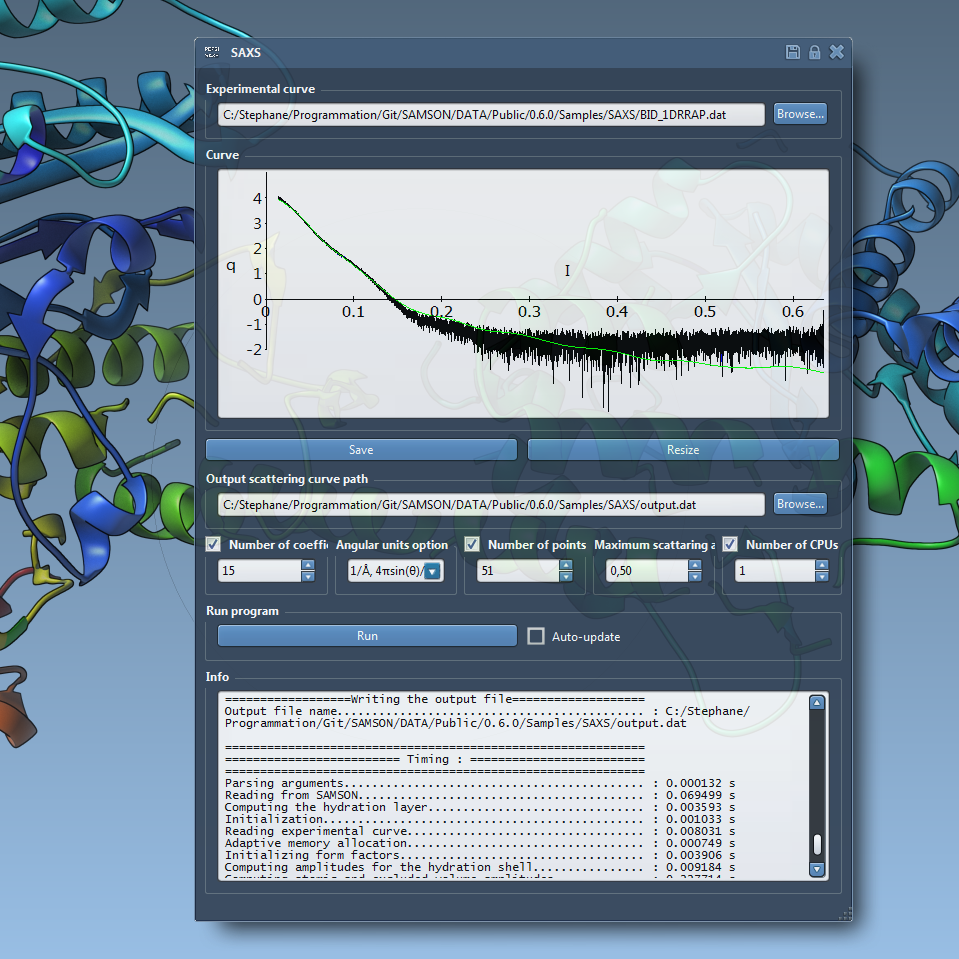
We show how to use the SAMSON app for analysis and fitting of Small-angle X-ray scattering curves in SAMSON. In particular, we show how the interactive update feature helps controlling the quality of the structure.
- 3:30 pm – 4:00 pm: Coffee break
- 4:00 pm – 4:30 pm: Docking ligands into proteins with AutoDock Vina - Show (all) - Hide (all)
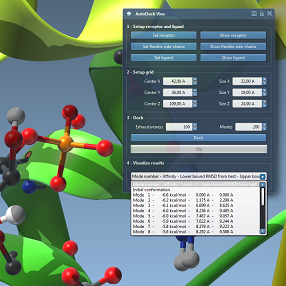
We demonstrate how to use the SAMSON app integrating AutoDock Vina from the Scripps institute. We show how to select the receptor, flexible chains and the ligand, as well as how to control the active degrees of freedom in the ligand. We show how to set up calculations and visualize results, as well as export the most promising conformations.
- 4:30 pm – 5:00 pm: Gaming for education - Show (all) - Hide (all)
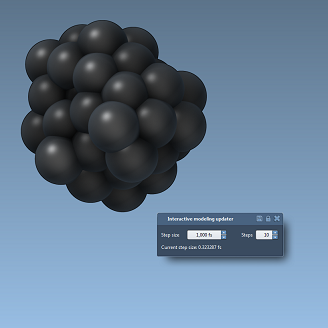
We demonstrate SAMSON apps created for high school students to explain various chemistry and physics notions, including temperature and crystal formation. In particular, we show how a group of particles subject to a Lennard-Jones force field and interactively minimized may serve as an introduction to the formation of molecules and materials.
- 7:00 pm: Dinner
Day 3 – Wednesday, June 21
- 7:30 am – 8:30 am: Breakfast
- 8:30 am – 9:00 am: Analyzing Transmission Electron Microscopy images of graphene - Show (all) - Hide (all)
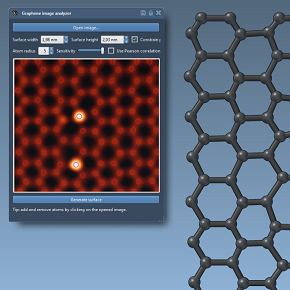
We show how to use a SAMSON app to analyze Transmission Electron Microscopy (TEM) images of graphene directly in SAMSON. In particular, we show how to set up the sensitivity of atom detection, and we demonstrate how a graphene structure may be created from the app based on the TEM image, to be exported and simulated.
- 9:00 am – 9:30 am: Generating crystal models - Show (all) - Hide (all)
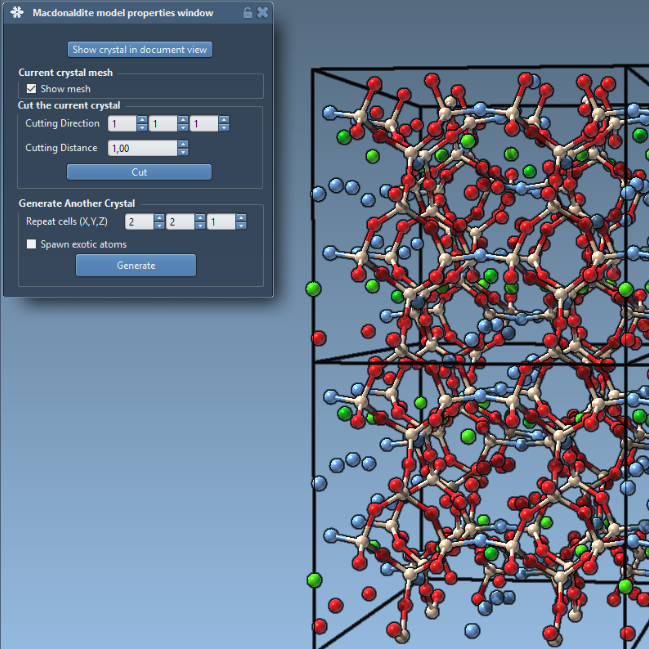
We show how to use the crystal generator app to easily create crystal models in SAMSON. We also show how to import cif models, visualize and edit them.
- 9:30 am – 10:00 am: Performing Orbital-Free Density Functional Theory calculations - Show (all) - Hide (all)
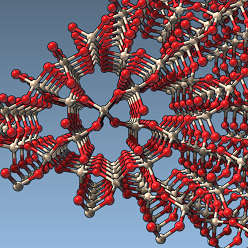
We demonstrate how to perform Orbital-Free Density Functional Theory (OF-DFT) calculations in SAMSON. In particular, we show how to set up the system and calculation, as well as combine these calculations with simulators in SAMSON to perform energy minimization.
- 10:00 am – 10:30 am: Coffee break
- 10:30 am – 11:00 am: Using SAMSON documents - Show (all) - Hide (all)
We discuss about how different versions of SAMSON and SAMSON Elements are managed, and how semantic versioning is used to control the interaction between SAMSON and SAMSON Elements. We discuss the SAMSON file formats (*.sam and *.samx), as well as automatic updates of SAMSON and SAMSON Elements.
- 11:00 am – 12:30 pm: Brainstorming – The SAMSON roadmap - Show (all) - Hide (all)
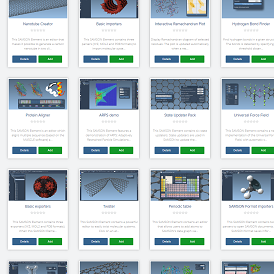
We review and conclude the Users program, and discuss about where SAMSON should head in upcoming versions. We brainstorm about possible new user features, and prioritize them.
- 12:30 pm – 2:00 pm: Lunch
End of the Users program
Beginning of the Developers program
- 10:30 am – 12:30pm: Arrival
- 12:30 pm – 2:00 pm: Lunch
- 2:00 pm – 3:30 pm: Welcome – Introduction and installation of the SAMSON SDK - Show (all) - Hide (all)
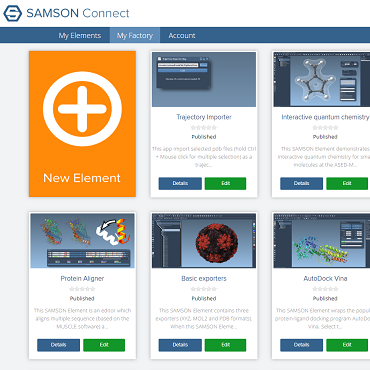
We introduce the Developers program of the SAMSON School, and the SAMSON Software Development Kit (SDK) that we will use to develop SAMSON Elements. We set up a programming environment for everyone, and we install the SAMSON SDK. We explore the SAMSON Connect platform from the developer point of view. We explain the structure of the SAMSON SDK. We introduce the libraries composing the SDK, and the main classes used to represent atoms, molecules and data graph nodes in SAMSON. We describe naming conventions, as well as the typical file structure of a SAMSON Element.
- 3:30 pm – 4:00 pm: Coffee break
- 4:00 pm – 5:00 pm: Programming new apps - Show (all) - Hide (all)
We use the SAMSON Element Generator to create the template of a new SAMSON app. We use this template to develop our first app: a tool to perturb atoms positions in the document. We explore the creation of the user interface, the retrieval of atoms in a document or in a user selection, and perturb atom positions using a random number generator. We use SAMSON's holding mechanism to let users undo and redo their actions.
- 5:00 pm – 5:30 pm: Editing and searching SAMSON documents - Show (all) - Hide (all)
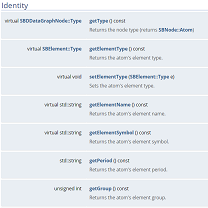
We show how to add and remove nodes from SAMSON documents, how to construct molecules using the SDK, and how to manipulate the SAMSON data graph with indexers and predicates, which power SAMSON's Node Specification Language. We explain how to construct and manipulate predicates, and how to use them to search data graph nodes.
- 5:30 pm – 6:00 pm: Programming Graphical User Interfaces with Qt - Show (all) - Hide (all)
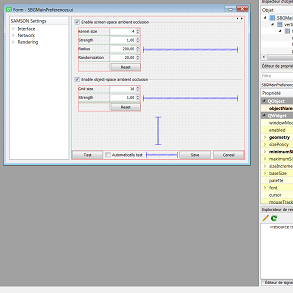
We further explore how to create user interfaces for SAMSON Elements using Qt and SAMSON windows. We introduce frequently used widgets, layouts, and explain Qt's signals and slots mechanism used to make interfaces react to user actions in more detail. We implement various interfaces to perform basic actions in SAMSON, and show how to save and load user-defined settings.
- 7:00 pm: Dinner
Day 4 – Thursday, June 22
- 7:30 am – 8:30 am: Breakfast
- 8:30 am – 9:00 am: Handling units and dimensional analysis - Show (all) - Hide (all)

In SAMSON, physical quantities are strongly typed. We show how to define and manipulate physical quantities and their units, in order to help ensure the quality of the calculations. We show how to convert between units, define new units and constants, and program an app that performs energy conversions.
- 9:00 am – 9:30 am: Importing and exporting data - Show (all) - Hide (all)
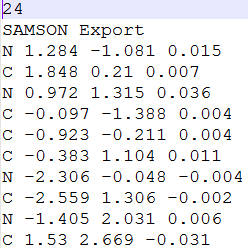
We describe how to import and export data into SAMSON. We implement an XYZ parser that creates a structural model based on atom coordinates and types, and we export an XYZ file based on the contents of the active document or the user selection.
- 9:30 am – 10:00 am: Creating novel visualizations - Show (all) - Hide (all)
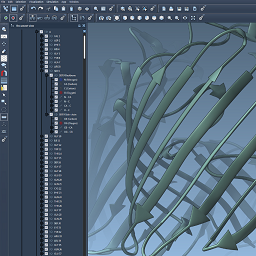
We introduce visual models, and show how they can be used to provide new visual representations to users. We introduce SAMSON's various display functions used to simplify the rendering of spheres, cylinders, surfaces, etc. We program a van der Waals representation of a group of atoms. We show how to make objects selectable by the user.
- 10:00 am – 10:30 am: Coffee break
- 10:30 am – 11:30 am: Developing new editors - Show (all) - Hide (all)
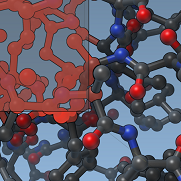
We demonstrate how to develop editors. We show how to react to mouse and keyboard events, and implement an editor that pushes atoms around. We show how to convert back and forth between screen coordinates and world coordinates, as well as pick data graph nodes in the viewport.
- 11:30 am – 12:30 am: Programming new force fields - Show (all) - Hide (all)
We explain how to program new force fields in SAMSON. We introduce dynamical models and interaction models, and show how to create and apply a set of springs to a group of atoms. We show how to compute energies and forces based on the information contained in the dynamical model.
- 12:30 pm – 2:00 pm: Lunch
- 2:00 pm – 2:30 pm: Programming new simulation methods - Show (all) - Hide (all)
We explain how to program new simulation methods in SAMSON. We introduce state updaters, the data graph nodes used to perform interactive simulation, and we implement a Metropolis Monte Carlo sampler. We show how to save the best structures found by the Monte Carlo minimizer in the document.
- 2:30 pm – 3:00 pm: Integrating external programs - Show (all) - Hide (all)
We explain how to develop a SAMSON app that integrates an external executable. In particular, we implement a prototype app that a) creates a configuration file on disk based on the parameters the user has entered in the app GUI, b) launches the external executable and c) retrieves the output of the external executable and imports it into SAMSON.
- 3:00 pm – 3:30 pm: Integrating web services - Show (all) - Hide (all)

We demonstrate how to connect SAMSON to a web service using Qt networking capabilities. In particular, we show how to fetch a protein structure from the protein databank, write a file to the disk, and import the structure into SAMSON.
- 3:30 pm – 4:00 pm: Coffee break
- 4:00 pm – 5:30 pm: Developing adaptive algorithms - Show (all) - Hide (all)
We introduce SAMSON's signals and slots mechanism that can be used to make SAMSON Elements react to events sent by atoms, bonds, models, etc., and develop adaptive algorithms. We develop an app that computes the center of mass of a group of atoms, and incrementally updates the center of mass when an atom moves.
- 5:30 pm – 6:00 pm: Saving and loading custom nodes - Show (all) - Hide (all)
We explain serialization, the mechanism that underlies various processes in SAMSON, used in particular to copy and paste data graph nodes, as well as save and load SAMSON documents. We show how to serialize and unserialize custom data graph nodes (e.g. new visual models that we want to save and share).
- 7:00 pm: Dinner
Day 5 – Friday, June 23
- 7:30 am – 8:30 am: Breakfast
- 8:30 am – 10:00 am: Sharing functionality between SAMSON Elements - Show (all) - Hide (all)

SAMSON's introspection mechanism makes it possible to share functionality between SAMSON Elements without having access to their source code. We explain the introspection mechanism, and we show how to expose, create and share classes between SAMSON Elements.
- 10:30 am – 11:00 am: Coffee break
- 10:30 am – 11:00 am: Distributing new SAMSON Elements on SAMSON Connect - Show (all) - Hide (all)
We explain how to use the SAMSON Element Packager to prepare a SAMSON Element for distribution on SAMSON Connect. We show how to upload screenshots and describe SAMSON Elements, manage collaborators allowed to edit the SAMSON Element, as well as manage versions of SAMSON Elements.
- 11:00 am – 12:30 pm: Brainstorming – The SAMSON SDK roadmap - Show (all) - Hide (all)
We review and conclude the Developers program, and discuss about where the SAMSON SDK should head in upcoming versions. We brainstorm about possible new developer features, and prioritize them.
- 12:30 pm – 2:00 pm: Lunch
- 2:00 pm: departure
To register, please visit this page.






